Developing proximity biotinylation assays in plants to expand the interactomics toolbox.
Proximity biotinylation uses a promiscuous biotin ligase, which causes biotinylation of proteins in the vicinity of the bait. These biotinylated proteins can be identified using mass spectrometry without the need to maintain the protein-protein interactions during the purification. This tool is therefore especially suited for interactions between cytosolic and transmembrane proteins. We are designing protocols to adequately perform proximity biotinylation in plants. This includes optimizing the temperature conditions, biotin concentration as well as the timing of biotin addition and the use of linker sequences between the ligase and the bait to achieve a bigger radius. We are also optimizing the detection of biotinylated peptides and we are developing methods to filter out false positives. This work runs in close collaboration with the functional interactomics group of Prof. Geert De Jaeger (PSB-VIB).
Arora et al., The Plant Cell 2020 PMID: 32843435
Cuadrado et al., JExBot 2024 PMID: 38437582
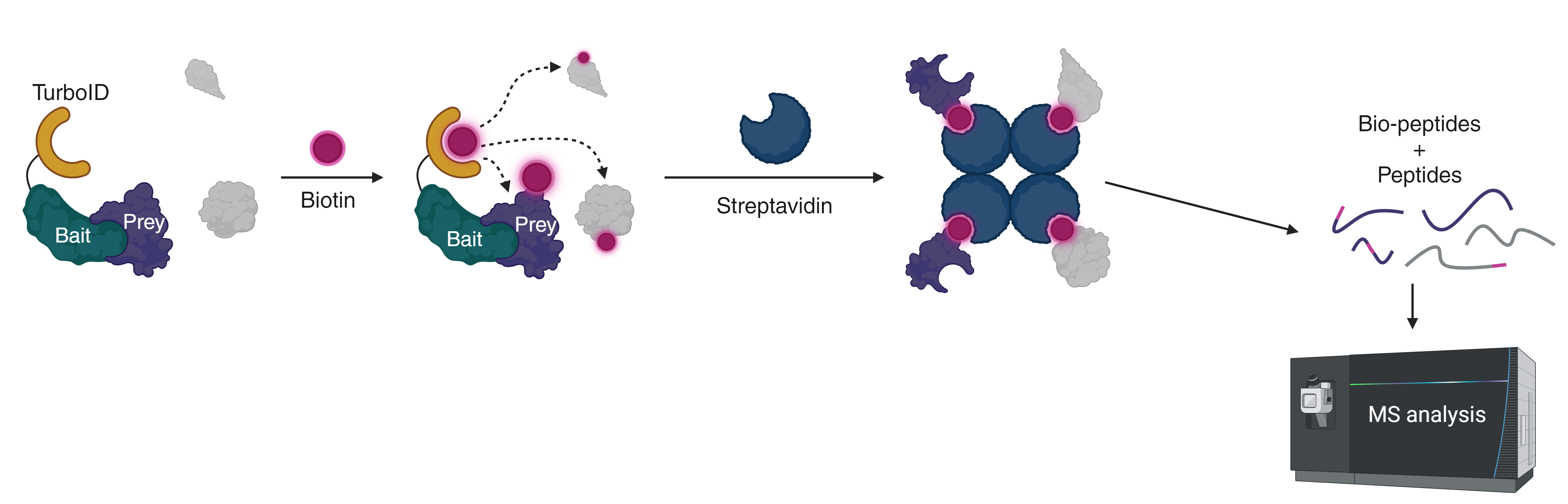
Schematic representation of proximity-biotinylation dependent interactomics. Interactors (prey) of the bait protein as well as proteins in the vicinity of the bait are labeled with biotin and captured on Streptavidin-coated beads. Following harsh washing steps, on bead trypsination and elution allows to identify both biotinylated as well as non-biotinylated peptides via mass-spectrometry analysis. Image adapted from (Cuadrado et al., 2024).