Visualizing protein-protein interactions in plants.
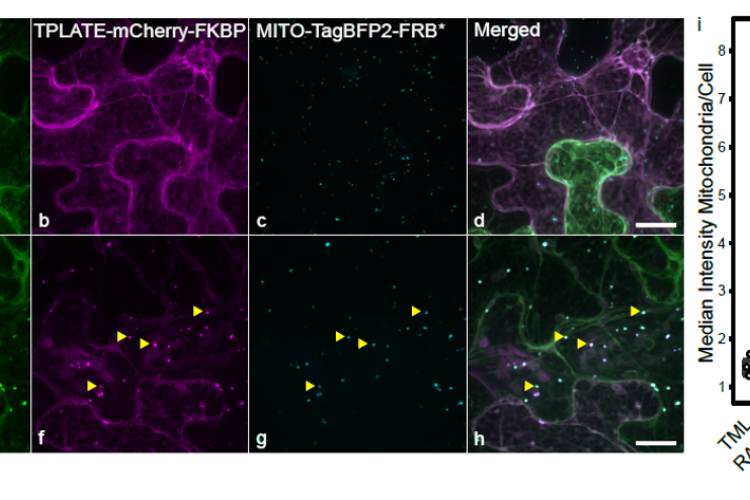
To gain insight into protein-protein interactions that occur in planta, we expanded the interaction assay toolkit. We have developed Knocksideways in plants (KSP) as an imaging-based protein-protein interaction assays to visualize binary as well as higher-order interactions by. KSP can be compared to an intracellular Co-IP experiment. The tool uses the ability of rapamycin to alter the localization of a bait protein and its interactors via the heterodimerization of FKBP and FRB domains. KSP is inherently free from many limitations, which other PPI systems hold. It is an in vivo in planta tool, it is flexible concerning the orientation of protein tagging as long as this does not interfere with the interaction and it is compatible with a broad range of fluorophores. KSP is also a conditional tool and therefore does not require additional controls.
We so far have generated anchor constructs targeting the FRB* domain to the plasma membrane, the mitochondria, the nucleus and the microtubule cytoskeleton. We have also developed scripts fro imageJ that allow to quantify the ineractions.
Next to transient expression in N. benthamiana, we have also shown that the system works in stable Arabidopsis lines.
Winkler et al., The Plant Cell 2021 PMID: 33793859
Dragwidge et al., Nature Cell Biology 2024 PMID: 38347182
Developing proximity biotinylation assays in plants to expand the interactomics toolbox.
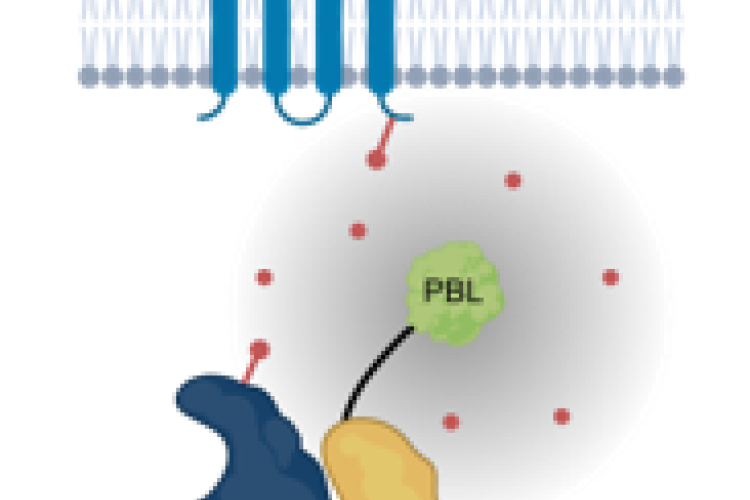
Proximity biotinylation uses a promiscuous biotin ligase, which causes biotinylation of proteins in the vicinity of the bait. These biotinylated proteins can be identified using mass spectrometry without the need to maintain the protein-protein interactions during the purification. This tool is therefore especially suited for interactions between cytosolic and transmembrane proteins. We are designing protocols to adequately perform proximity biotinylation in plants. This includes optimizing the temperature conditions, biotin concentration as well as the timing of biotin addition and the use of linker sequences between the ligase and the bait to achieve a bigger radius. We are also optimizing the detection of biotinylated peptides and we are developing methods to filter out false positives. This work runs in close collaboration with the functional interactomics group of Prof. Geert De Jaeger (PSB-VIB).
Arora et al., The Plant Cell 2020 PMID: 32843435
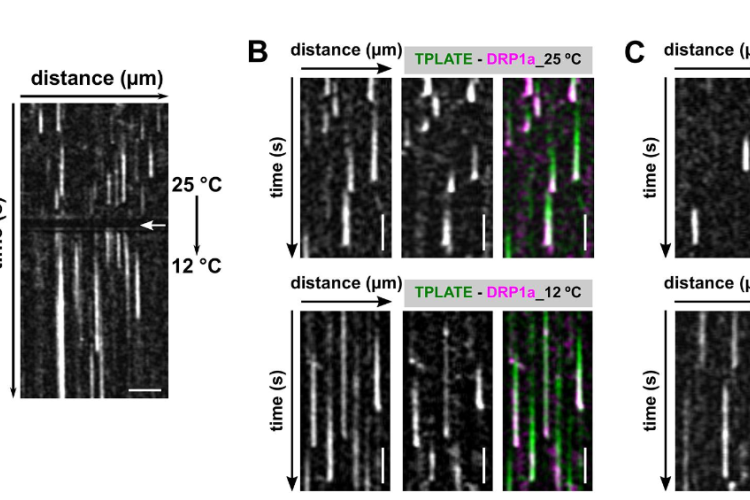
Temperature-modulated live cell imaging.
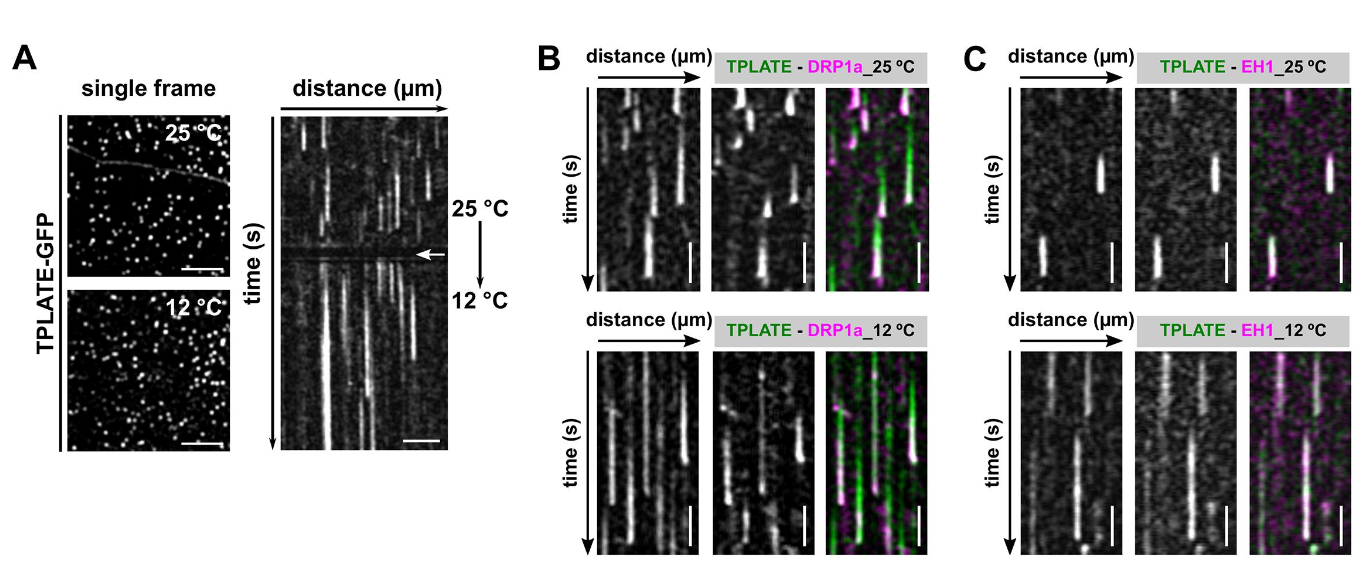
Dynamic imaging of fast-occurring processes is hampered by the fact that the signal/noise ratio of a fluorescent image is inversely related to the exposure time. The exposure time in turn limits the imaging speed and thereby the temporal resolution. We are increasing the temporal resolution of live-cell imaging plant samples by modulating the temperature of the samples in situ on the microscope stage as lowering the temperature slows down biological processes.
CME is a fast and stepwise process and involves the sequential recruitment of several players. Adaptor complexes, the clathrin scaffold and endocytic accessory proteins all arrive and depart at different time points during the process. To visualize the temporal dynamics of the various players, we are generating fluorescently tagged functional fusions of TPC subunits and are performing dual color spinning disc microscopy on complemented double mutant lines at ambient and at lowered temperature. Imaging endocytosis at lowered temperature delays the process and allowed us to increase the temporal resolution by which the effector proteins are recruited to the plasma membrane.
Next to this, we use our temperature-modulatory setup also to study the effect of high temperature stress on plants. Here we combine live-cell imaging of fluorescently tagged proteins with temperature changes to observe how this alters their localization and properties.